Elham Azizi, PhD
- Herbert & Florence Irving Assistant Professor of Cancer Data Research in the Irving Institute for Cancer Dynamics
- Assistant Professor of Biomedical Engineering
On the web

Overview
Academic Appointments
- Herbert & Florence Irving Assistant Professor of Cancer Data Research in the Irving Institute for Cancer Dynamics
- Assistant Professor of Biomedical Engineering
Gender
- Female
Credentials & Experience
Education & Training
- PhD, Boston University, Bioinformatics
- MS, Boston University, Electrical Engineering
- BS, Sharif University of Technology, Tehran, Iran.
Honors & Awards
- Early-Career Innovator in Science Award in Cancer Immunology, Takeda and the New York Academy of Sciences (one awardee across the globe), 2024.
- Allen Distinguished Investigator Award, Allen Institute, 2023.
- NHGRI Award for Supporting Talented Early Career Researchers in Genomics (R01), 2023.
- CZI Science Diversity Leadership Award, Chan Zuckerberg Initiative and the National Academies of Sciences,
Engineering, and Medicine, 2022. - NSF CAREER Award, 2022.
- Provost’s Grant for junior faculty contributing to the diversity goals of Columbia University, 2022.
- Columbia Research Initiatives in Science & Engineering (RISE) Award, 2021.
- Irving Endowed Assistant Professorship in Cancer Data Research, Columbia University, 2020.
- Tri-Institutional Breakout Prize for Junior Investigators, Weill Cornell Medicine, Rockefeller University, and Memorial Sloan Kettering Cancer Center, 2019.
- Next Generation in Biomedicine (20 scientists selected worldwide), Broad institute of MIT and Harvard, 2018.
- NIH NCI Pathway to Independence Award (K99/R00), 2018.
- American Cancer Society Postdoctoral Fellowship, 2017.
Research
Dr. Azizi's work aims to characterize the complex populations of interacting cell types in the tumor microenvironment and their underlying circuitry by leveraging cutting-edge single-cell genomic technologies. Her work involves developing novel machine learning methods to address the statistical and computational challenges inherent to analyzing data from single-cell technologies. By integrating single-cell and multi-omics data using probabilistic modeling approaches, she aims to infer dysregulated programs driving cancer stem cells as well as the reprogramming of immune cells leading to immune dysfunction. These interpretable models can ultimately help guide improved and personalized cancer therapies.
Research Interests: Cancer Immunology, Computational Biology, Genomics
Grants
- R21HG012639 PI: Azizi 1.7 months Total: $435,475 NIH/NHGRI 9/21/2023-8/30/2025 Title: Computational toolbox for spatial transcriptomic analysis of complex tissues
- R01HG012875 PI: Azizi 2.4 months Total: $2,219,971 NIH/NHGRI 02/17/2023 - 12/31/2027 Title: Machine learning methods for interpreting spatial multi-omics data
- Allen Distinguished Investigator Award PI: Simunovic, McFaline, Azizi 0.36 months Total: $1,500,000 The Paul G. Allen Frontiers Group 12/01/2023-12/30/2026 Title: Sex hormone morphogenesis: a new frontier in studying organ development
- Science Diversity Leadership Award PI: Azizi 2.0 months Total: $1,150,000 Chan Zuckerberg Institute 12/01/2022 - 11/30/2027
- CAREER CBET 2144542 PI: Azizi 0.25 months Total: $500,030 NSF 2/15/2022-1/31/2027 Title: CAREER: Integrative modeling of intercellular interactions in the tumor microenvironment
- R01CA266446 PI: Izar; Co-I: Azizi 0.36 months Total: $2,453,430 NIH/NCI 9/1/2022-8/31/2027 Title: The role of the CD58:CD2 axis in cancer immune evasion and resistance to immunotherapy
- SCOR-22937-22 PI: Soiffer, Wu; Co-I: Azizi 0.36 months Total: $5,000,000 Leukemia & Lymphoma Society 10/1/2022 – 9/20/2027 Title: Understanding and Overcoming Mechanisms of Immune Evasion after Allogeneic Transplant
Selected Publications
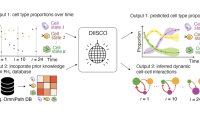
- Park C*, Mani S*, Beltran-Velez N, Maurer K, Gohil S, Li S, Huang T, Knowles DA, Wu CJ, Azizi E^. A Bayesian framework for inferring dynamic intercellular interactions from time-series single-cell data. Genome Research. 2024.
- Gu J, Iyer A, Wesley B, Taglialatela A, Leuzzi G, Hangai S, Decker A, Gu R, Klickstein N, Shuai Y, Jankovic K, Parker-Burns L, Jin Y, Zhang JY, Hong J, Niu S, Chou J, Landau DA, Azizi E, Chan EM, Ciccia A, Gaublomme JT. CRISPRmap: Sequencing-free optical pooled screens mapping multi-omic phenotypes in cells and tissue. bioRxiv. 2023:2023-12. Nature Biotechnology. 2024.
- He S*, Jin Y*, Nazaret A*, Shi L, Chen X, Rampersaud R, Dhillon BS, Valdez I, Friend LE, Fan JL, Park CY, Mintz Y-H, Carrera D, Fang KW, Mehdi K, Rohde M, McFaline-Figueroa JL, Blei D, Leong KW, Rudensky AY^, Plitas G^, Azizi E^. Starfysh integrates spatial transcriptomic and histologic data to reveal heterogeneous tumor–immune hubs. Nature Biotechnology, 2024.
- Fuller J, Abramov A, Mullin D, Beck J, Lemaitre P, Azizi E^. A Deep Learning Framework for Predicting Patient Decannulation on Extracorporeal Membrane Oxygenators: Development and Model Analysis Study. JMIR Biomedical Engineering. 2024.
- Liu Y, Jin Y, Azizi E^, Blumberg AJ^. CellStitch: 3D Cellular Anisotropic Image Segmentation via Optimal Transport. BMC Bioinformatics. 24, 480. 2023.